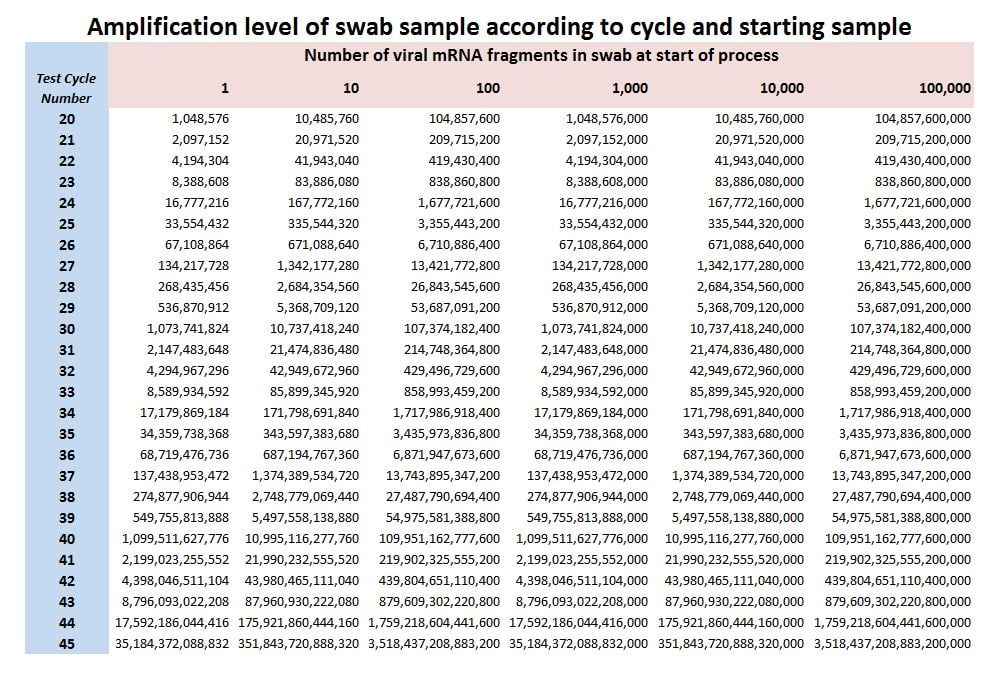
RT-PCR Test Amplification Table
from John Dee’s Almanac excellent facebook site – John Dee’s Almanac | Facebook
His own words below
We’ve had some excellent discussion on this in the group recently so I fancied knocking this table up whilst my tea brews. If you start with just 1 viral mRNA chain and flip this into 1 DNA analog using Reverse Transcriptase, then run Polymer Chain Reaction you’ll theoretically end up with 1,048,576 DNA chains after 20 cycles, this being the number in the top left of the table. Progression from there is yet more doubling. But cycle number is only half of the issue! Other things we need to consider are A.) How big our swab sample is in the first instance and B.) How many viral mRNA fragments our sample contains. No doubt there are other tricky issues. A jolly good poke up the nose means we may start with a large sample compared to a wimpy wipe. It should be obvious from this table that non-standard nose-poking alone will lead to different test results and then, of course, there is the matter of genuine viral loading. At 45 cycles we’ve managed to get 1 mRNA fragment amplified to 35,184,372,088,832 DNA chains – a number that surpasses the 26,843,545,600,000 DNA chains obtained from just 28 cycles based on a swab containing 100,000 viral mRNA fragments. One of these makes clinical sense, the other does not. As Professor Heneghan of the Centre for Evidence-Based Medicine at Oxford University said of PCR early last year, we have no idea whether 1 viral mRNA fragment has any clinical meaning but, hey ho, a cycling we will go!